Project Documentation
Project Title: BiœmuS: A New Tool for Neurological Disorders Studies
Problem Statement:
Millions of people globally suffer from neurological disorders that significantly impair cognitive and motor functions. Current pharmacological treatments are often limited to managing symptoms rather than providing a cure. There is a pressing need for alternative therapeutic approaches, including neuroprostheses, which require a deep understanding of the interactions between biological neural networks and artificial systems. The challenge is to develop a tool that can facilitate real-time emulation and hybridization experiments to advance neuromorphic engineering and neuroprosthetic technologies.
Objectives:
- Develop a Biomimetic Spiking Neural Network (SNN):
- Create a low-cost, user-friendly SNN platform capable of real-time emulation for biohybrid experiments.
- Ensure the system can emulate biophysically detailed neural network dynamics accurately.
- Facilitate Biohybrid Experiments:
- Enable seamless interaction between biological cells and the artificial neural network.
- Provide standard biophysical interfaces for integrating various biological cells into experiments.
- Enhance Neuroprosthetic Development:
- Contribute to the creation of neuromorphic-based neuroprostheses for bioelectrical therapeutics.
- Ensure the platform supports adaptive stimulation at biological timescales to improve communication with biological networks.
- Improve Accessibility and Usability:
- Prioritize cost-efficiency and flexibility to make the platform accessible to a wide range of researchers and clinicians.
- Develop intuitive software for easy configuration, monitoring, and control of the SNN.
- Promote Real-World Applications:
- Demonstrate the practicality and potential of the platform for real-world applications in neurological disorder treatment.
- Showcase the system's capability in both in vitro and in vivo experimental models to validate its effectiveness and versatility.
By achieving these objectives, the project aims to bridge the gap between biological and artificial neural systems, paving the way for advanced treatments for neurological disorders through innovative neuromorphic and neuroprosthetic technologies.
Overview
BiœmuS is an advanced system designed for the study and treatment of neurological disorders through real-time emulation and hybridization using a biomimetic Spiking Neural Network (SNN). The system is developed to facilitate the investigation and replication of neural network dynamics, bridging the gap between biological and artificial neural networks.
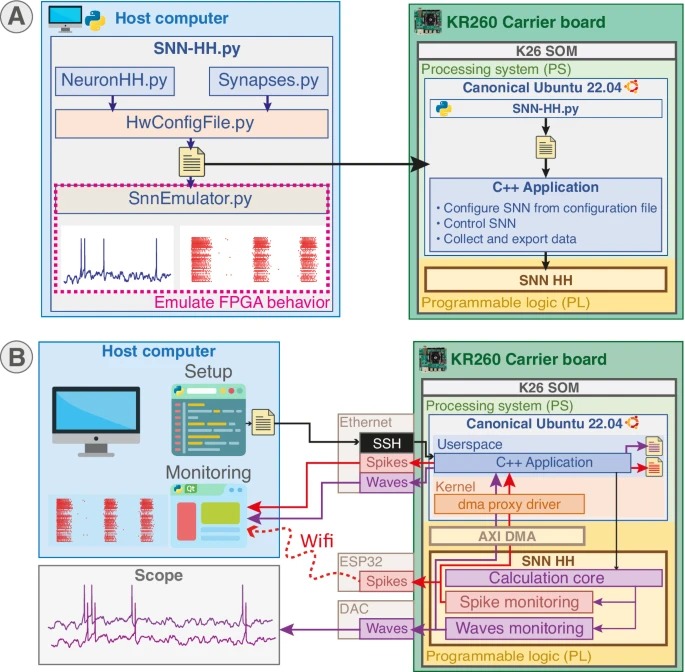
Overview of the system architecture 📷
Network configuration file 📷
Monitoring channels 📷
Importance
The BiœmuS project addresses critical needs in the treatment and understanding of neurological disorders, which affect millions globally. Traditional pharmacological treatments are often limited in their effectiveness, primarily managing symptoms rather than providing cures. BiœmuS offers a promising alternative through its neuromorphic approach, enhancing the development of neuroprostheses and electroceutical treatments.
Applications
BiœmuS has several key applications:
- Neurological Disorder Studies: It allows detailed studies of neural dynamics, aiding in understanding brain functions and pathologies.
- Neuroprostheses Development: Facilitates the creation of advanced neuroprostheses that mimic biological neural activities.
- Real-time Emulation: Supports real-time emulation of neural networks for biohybrid experiments, enhancing research in neuromorphic engineering.
- Electroceutical Treatments: Provides a platform for developing electroceutical therapies for diseases like Parkinson’s and epilepsy.
- Brain-Computer Interfaces (BCIs): Enhances BCI research by providing real-time adjustments to improve system performance and user interaction.
Hardware Requirements
- Kria KR260 Carrier Board: Integrates the hardware components required for the project.
Software Requirements
- Canonical Ubuntu 22.04: Operating system running on the processing system.
- Python 3.x: For running configuration scripts.
- C++ Compiler: For compiling control applications.
- ZeroMQ: Networking library for data communication.
Using Kria KR260
The Kria KR260 board is crucial for the BiœmuS project as it integrates both programmable logic (FPGA) and processors, providing a versatile and powerful platform for running the Spiking Neural Network. The FPGA handles real-time neural network computations, while the processors manage configuration, monitoring, and control.
Features of Kria KR260:
- Real-time Emulation: The FPGA enables precise, low-latency computations required for real-time emulation of neural networks.
- Versatile Communication: Multiple communication interfaces (Ethernet, Wi-Fi, PMODs) ensure seamless integration with various external devices and networks.
- Ease of Use: Running a standard Ubuntu operating system simplifies software development and integration, making the system accessible to neuroscientists and engineers.
Get Started
Setup Ubuntu operating system
This steps prepares the operating system to run BioemuS on KR260 or KV260 targets.
- Download Canonical Ubuntu image for AMD target from here
- Ubuntu 20.04
- Ubuntu 22.04 (:warning:KV260: not supported from stock, boot firmware update required)
Official tutorials
- Kria™ KR260 Getting Started Guide for Ubuntu 22.04
- Kria™ KV260 Getting Started Guide for Ubuntu 22.04
- Flash image on SD card
- Insert SD card
- Connect USB serial (1) to PC (USB micro B)
- USB to UART Bridge: Interface 0, 115200 baud
- Connect Ethernet (2) to PC or local network
- SSH connection: @username ubuntu @port 22
- Connect power supply (3)
- Login to Ubuntu
# Default
login: ubuntu
password: ubuntu
# bioemus
login: ubuntu
password: kriachan- Setup internet access
(1) Connected to local network
(2) Shared over Ethernet
Internet connection can be checked using nmcli general status
% nmcli general status
STATE CONNECTIVITY WIFI-HW WIFI WWAN-HW WWAN
connected full enabled enabled enabled enabled- Get the IP assigned to the board using ifconfig command (inet value of eth1, here 192.168.137.248)
ubuntu@kria:~$ ifconfig
eth0: flags=4099<UP,BROADCAST,MULTICAST> mtu 1500
ether 00:0a:35:0f:d6:48 txqueuelen 1000 (Ethernet)
RX packets 0 bytes 0 (0.0 B)
RX errors 0 dropped 0 overruns 0 frame 0
TX packets 0 bytes 0 (0.0 B)
TX errors 0 dropped 0 overruns 0 carrier 0 collisions 0
device interrupt 38
eth1: flags=4163<UP,BROADCAST,RUNNING,MULTICAST> mtu 1500
inet 192.168.137.248 netmask 255.255.255.0 broadcast 192.168.137.255
inet6 fe80::e83c:990d:d8ba:55e7 prefixlen 64 scopeid 0x20<link>
ether 00:0a:35:0f:e0:79 txqueuelen 1000 (Ethernet)
RX packets 109 bytes 17011 (17.0 KB)
RX errors 0 dropped 0 overruns 0 frame 0
TX packets 96 bytes 9737 (9.7 KB)
TX errors 0 dropped 0 overruns 0 carrier 0 collisions 0
device interrupt 37
lo: flags=73<UP,LOOPBACK,RUNNING> mtu 65536
inet 127.0.0.1 netmask 255.0.0.0
inet6 ::1 prefixlen 128 scopeid 0x10<host>
loop txqueuelen 1000 (Local Loopback)
RX packets 274 bytes 21577 (21.5 KB)
RX errors 0 dropped 0 overruns 0 frame 0
TX packets 274 bytes 21577 (21.5 KB)
TX errors 0 dropped 0 overruns 0 carrier 0 collisions 0eth0: bottom PS Ethernet port eth1: top PS Ethernet port
- Start SSH session at the IP address (MobaXterm, or console if Linux)
# MobaXterm
Remote host: 192.168.137.248
Username: ubuntu
Port: 22
# Linux
ssh ubuntu@192.168.137.248- Update system
sudo apt-get update && sudo apt upgrade
sudo reboot nowOn Kria™ KR260 the boot time can be quite long (~180 seconds) from the TPM self test message.
- Install Xilinx Development Tools package
# Ubuntu 20.04
sudo snap install xlnx-config --classic --channel=1.x
# Ubuntu 22.04
sudo snap install xlnx-config --classic --channel=2.x
# Initialize xilinx config package
xlnx-config.sysinit
# Bootgen
sudo apt install bootgen-xlnx- Install BioemuS dependencies
# ZeroMQ
sudo apt install libzmq3-dev
# Python
sudo apt install python3-pip python3-pyqt5- (optional, install Ubuntu Software)
sudo snap install snap-store- (optional, utilities)
sudo apt install net-tools
sudo apt install devmem2- (optional, disable GUI for remote use)
# Disable GUI
sudo systemctl set-default multi-user
# Enable GUI
sudo systemctl set-default graphicalSetup Bioemus
This procedure "installs" Bioemus on the board by building the different requirements for Bioemus to run.
- Copy sw/target/kria on the board at ~/ or /home/ubuntu (i.e. using SFTP with MobaXterm, USB drive, ...) or clone from github.
git clone https://github.com/Ceramic-Blue-Tim/bioemus.git gitbioemus
cp -r gitbioemus/sw/target/kria bioemus- Rename folder to bioemus
mv kria bioemus- Run initialization
cd bioemus
chmod +x init.sh
source ./init.sh
echo "source ${BIOEMUS_PATH}/init.sh" >> ~/.bashrcIn case of scripts not running, try converting all .sh to unix format using dos2unix (sudo apt install dos2unix then dos2unix *.sh)
- Build all recipes (software: C++ application, firmware: device tree and bitstream platform, drivers: DMA proxy driver)
./build.shThe different depedencies can be build independently by passing the arguments software|firmware|drivers.
Generate configuration files
This step details the generation of the configuration files of the system performed on the host computer. There are two configuration files: one to configure the C++ application (.json) and one to configure the SNN (.txt). Both files are generated by the Python but only the .json can be modified after generation.
Detailed procedure to generate configuration file can be found here.
Create python environment using conda
- Create the environment
conda create -n bioemus python=3.11- Install packages required
conda activate bioemus
pip install -r host/requirements.txtGenerate configuration files from notebook
- Open the host folder in VSCode and open the file main.ipynb
- Run the Generate configuration file section
- Default path for configuration files is host/export/
The parameters have to be described in the file gen_config.py
Transfer configuration files to the board
- Transfer configuration files (.json and .txt) on the board in the folder bioemus/config/
Run Bioemus
Application settings
- Application settings can be modified from the JSON configuration file
{
"fpath_hwconfig": "/home/ubuntu/bioemus/config/hwconfig_stim.txt",
"emulation_time_s": 10,
"sel_nrn_vmem_dac": [0, 1, 2, 3, 4, 5, 6, 7],
"sel_nrn_vmem_dma": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15],
"save_local_spikes": false,
"save_local_vmem": false,
"save_path": "/home/ubuntu/bioemus/data/",
"en_zmq_spikes": true,
"en_zmq_vmem": false,
"en_zmq_stim": true,
"en_wifi_spikes": false,
"ip_zmq_spikes": "tcp://*:5557",
"ip_zmq_vmem": "tcp://*:5558",
"ip_zmq_stim": "tcp://192.168.137.1:5559",
"bin_fmt_save_spikes": false,
"bin_fmt_save_vmem": false,
"bin_fmt_send_spikes": false,
"bin_fmt_send_vmem": false,
"nb_tstamp_per_spk_transfer": 100,
"nb_tstep_per_vmem_transfer": 190,
"en_stim": false,
"stim_delay_ms": 5,
"stim_duration_ms": 20
}- Example: saving local spikes and external stimulation
{
"save_local_spikes": true,
"save_path": "/home/ubuntu/bioemus/data/",
"en_zmq_stim": true,
"ip_zmq_stim": "tcp://192.168.137.1:5559",
"nb_tstamp_per_spk_transfer": 100,
}- Example: stimulation step and store local waves
{
"sel_nrn_vmem_dma": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15],
"save_local_vmem": true,
"save_path": "/home/ubuntu/bioemus/data/",
"nb_tstep_per_vmem_transfer": 190,
"en_stim": true,
"stim_delay_ms": 5,
"stim_duration_ms": 20
}
Run application
- Run application
./launch_app.sh <path_to_swconfig_file_json>Monitor using ZeroMQ
- Either run run_spkmon.py or the spike monitoring section from Monitoring in main.ipynb.
- Set the IP address of the board and port for spikes (5557)
tcp://192.168.137.248:5557- Press the connect button
· /!\ NB_FRAME_PER_BUFFER in monitoring/spkmon/spkmon/settings/config.py must be identical to "nb_tstamp_per_spk_transfer" (default 100)
· ZeroMQ sending has to be enabled in the .json file
· Smaller window width limits GUI freezing
Conclusion
BiœmuS leverages advanced neuromorphic engineering to provide a powerful tool for neurological research and treatment development. Its integration with the Kria KR260 board enhances its real-time emulation capabilities, making it a versatile and practical solution for both in vitro and in vivo studies. Through its applications in neuroprostheses, BCIs, and electroceutical treatments, BiœmuS stands to significantly impact the field of neurological disorder treatment.





Comments