The very first step in tackling a disease is the diagnosis, and its a hard problem. SARS-COV-2 outbreak is unusual as such a high percentage of people infected are asymptomatic and spread the disease whilst appearing healthy. Even for those people who will go on to get sick, COVID-19 does not always give reliable symptoms during its onset. The best way to slow this disease without inflicting huge economic pain is to quarantine only those people with the disease, but this needs a massive amount of testing.
A SolutionTesting centers are expensive to set up and require highly trained staff to run the complex devices needed to detect COVID-19. Another option is to do distributed testing using inexpensive devices. In areas of the world with poor transport infrastructure, this might be the only feasible option.
If we had a way to perform a biophysical assay that could determine if COVID-19 was present in a sample, in a device that was cheap enough to get to all corners of the globe, we might have a chance to slow the spread of the disease until a vaccine is developed.
This project will document how to cheaply and rapidly build an Arduino-based device that can detect a handful of COVID-19 viral particles from a nasal swab, and yet be easy and reliable enough to be used by untrained people in rough conditions. It will cost less than $30 to build and can be used thousands of times, with tests costing just a few cents each.
This project combines cutting-edge techniques in Molecular Biology for detecting COVID-19, with an automated Fluorescence spectroscopy system based on parts found in most Makerspaces.
Background Biology
All living things (and let's include viruses here), at the barest minimum, need both a body and a way to reproduce. Viruses are at the logical boundaries of what we can consider 'life' because they don't actually have much more of a body than a simple shell around their DNA or RNA. They don't need to eat, sleep, or even have a metabolism. All they need to do is infect a cell, and once inside, they hijack that cells' molecular machinery to produce new viral particles.
The 'shell' or capsid of a COVID-19 virus is made up of proteins and is covered in Spike proteins, which help the virus invade cells. This particular kind of virus is a positive-sense single-stranded RNA virus, which is different from the way we store our own molecular information, which is encoded in double-stranded DNA.
When someone is infected with COVID-19, their bodies try and mount a defense. The immune system learns to recognize the foreign proteins in the viral capsid and develops antibodies that can bind and attack the virus. But it is a race against time. The immune system has to learn about, develop, and build antibodies against the virus before the virus spreads and kills the too many cells it infects. The immune systems can also be provoked into over-responding and causing damage itself.
To make an accurate diagnosis for this disease, we can either:
- Detect the virus's proteins
- Detect the virus's RNA
- Detect the antibodies a patient generates against the virus over the course of the disease.
Let's look at each of these options: Detecting the viral proteins can very hard to do reliably, as there might be so few viral particles in the sample that it's nearly impossible to detect (and truly impossible to do cheaply), even if the patient is heavily infected. Detecting the patient's antibodies is a very reliable indicator, but only works well into the infection. Earlier on, the patient might still be very infectious, but the test would be negative as their bodies haven't yet developed the necessary antibodies.
RNA detection is the best option for early detection, as there are reliable methods to make huge numbers of copies of the viral RNA so that even a few viral particles can be reliably detected.
Background Molecular Biology
Being able to make copies of DNA unleashed the molecular biology revolution. The key innovation was the development of the Polymerase Chain Reaction or PCR. It allows us to make billions of copies of a piece of DNA quickly and cheaply, allowing us to clone DNA, trace our ancestors, and produce genetically modified organisms.
Want an analogy on how it works? Imagine a magical photocopier. If you want any section of a book copied, you just write the first and last sentence on pieces of paper and feed them into the magical machine along with a copy of the book. The machine makes billions of copies of the section you wanted, and the whole process cost just a few cents! That's how PCR works. You can easily synthesize little fragments of DNA (called primers), and used those to select which section of DNA you want to be copied. The magical copier machine is the DNA Polymerase, and you have versions of that nano-machine in every cell in your body.
The PCR process is conceptually simple: Each thermal cycle we a) Denature: We heat up our sample so high they the DNA double strands break apart, b) Anneal: We cool down the sample until a small piece of synthetics DNA (which is cheap and easy to manufacture by the billions) sticks to the single stand. Its base pair are designed to match only one exact sequence (think regex find string). c) Elongate: a special protein called a "Polymerase" functions as a nano-machine, grabbing nucleotides out of the liquid around it, and using them as building blocks that can make new DNA extending from the short synthetic DNA, using the single strand as a temple, in effect making a copy. As the new copy can itself act as a template, each round of this temperature cycling doubles the number of copies, and its usual to do 30 such rounds!
PCR can also be used to detect disease. This is because the DNA copying process only works if the primers are an exact match for the viral DNA! If we know a sequence of DNA that is only found in a certain virus (that's the job of the scientists), we can design very specific primers to only amplify DNA from that and only that virus!
The most commonly used detection method for SARS-COV-2 so far has been reverse transcription-polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR). It converts the RNA to DNA (reverse transcription), then uses PCR to try and make copies of a virus-specific sequence that might be present in the sample. If we measure the DNA amplification against the number of PCR cycles, we can estimate how much initial viral RNA was present in the sample, and so be able to estimate the progress of the disease.
However, RT-PCR requires expensive thermocycler devices that start in the thousands of dollars range and massively increase in price for qRT-PCR devices.
For a distributed and cheap solution, this simply can't work.
There is another method though, that's both much more complex on the biophysical side, but much, much more simple to perform practically. That method is called Loop-mediated isothermal amplification (LAMP) and is a single-tube technique for the amplification of DNA. It is a low-cost alternative to detect certain diseases, including SARS-COV-2, when LAMP is combined with a reverse transcription step to allow the detection of RNA (RT-LAMP).
LAMP is very difficult to explain in diagrams, but there are many videos that describe how it works. Basically though, instead of a normal using a PCR-type Polymerase, we use a type of polymerase that has 2 special properties:
- It has "reverse-transcription" activity, and can "read" RNA and "write" DNA. That means it can use RNA as a template to generate new and matching DNA strands.
- It has a "strand-displacement" activity. This means the Polymerase can actively "unzip" double-strand DNA to access the data inside, so we don't have to do a high-temperature Denaturation step, as with PCR.
Not only that but as the process requires 4 or 6 primers, it is actually way more specific than PCR, as all the sections need to match the template DNA!
What does all this mean?
Using the LAMP technique, we only need to mix the RNA from a few virus particles with a mixture of cheap chemicals, and hold it's at a steady temperature to produce huge amounts of DNA!
How much is a 'huge amount' of DNA? We're talking in the range of about a 10-billion-fold increase! Not bad for a reaction that costs cents!
But how do we detect that the DNA is produced? Luckily, there are a few colorimetric methods (tests that change color!) available, and the easiest of with a special dye called calcein. The reason this is such a great method it that we can accurately track the amount of DNA produced over time, as its directly proportional to the fluorescence produced. That means, like RT-PCR, we can track the progression of the reaction and determine how much virus was in the original sample.
I bought a 10-gram bag from eBay for a bit over 20 Euro. How much does that get me? Well, the molecular weight is 666.50 g/mol, and we need a final concentration of 20 mM Calcein to detect the reaction. That means we use:
Rearranging and filling in the numbers, we find we have enough calcein to make 750 milliliters of the reaction solution. As we only 25 microliters per reaction, my 20 euros has bought enough material for about thirty-seven thousand COVID19 tests! Or a bit over $0.0005-worth per test.
Let's not skip too many details, and look into the mechanism of how Calcein acts as a colorimetric reagent. Our reaction mixture contains DNTP's, or the individual base-pair sub-units that will be used to grow the newly forming DNA chains. Each 'letter' has three phosphate ground attached to it, which acts as the energy-supply to drive the reaction. One of the phosphates goes on the make up the 'backbone' of each DNA strand, and the other two are thrown away.
In our reaction mixture are two types of metal ions Magnesium and Manganese. The calcein dye is quenched by manganese (meaning that the metal ion sticks to and messes up the calcein's ability to fluoresce). As the reaction goes on, those 'waste' pyrophosphates build-up, and the manganese ions prefer to stick to the pyrophosphate than the calcein. This means the calcein now starts to build up calcein–magnesium complexes, that are visible as bright green fluorescence when under blue or UV light.
Although you can sometimes see the activated calcein by eye without UV, is we be better to accurately track the amount of fluorescence. That is because like RT-PCR described above if we track when we first see the fluorescence appear, we can calculate how many viral particles were in the original sample!
In the end, if there are any viral particles in the sample, the exponential creation of DNA will use up all the DNTP's, and every molecule of calcein will become fluorescent. But the more viral particle you start with, the sooner this will become measurable.
LAMP requires primers that bind to the transcripted viral DNA. Sets of primers we can use are https://www.biorxiv.org/content/10.1101/2020.04.21.052530v1.full:
Primer Sequences (5′-3′)
GeneN-F3 GCC AAA AGG CTT CTA CGC A
GeneN-B3 TTT GGC CTT GTT GTT GTT GG
GeneN-FIP TCC CCT ACT GCT GCC TGG AGT TTT CGG CAG TCA AGC CTC TTC
GeneN-BIP TCC TGC TAG AAT GGC TGG CAA TTT TTT TTG CTC TCA AGC TGG TTC A
GeneN-LoopF CGA CTA CGT GAT GAG GAA CGA
GeneN-LoopB GCG GTG ATG CTG CTC T
These primers can be orders online and usually come in 25 nanomole batches. That's not much material, but it's enough for many tests. If you order in bigger batches, it gets much cheaper.
To calculate it out, we need a concentration of primers of roughly 1 uM
M=n/V, where M is the concentration, n is the number of moles and V is the volume in liters.
That means the volume we can make is n/M, or 0.000000025 moles / 0.000001 micromolar, so we can calculate we can make 0.25 liters of reaction mixture. As we only need 50 microliters per reaction, that's about 500 reactions worths for about $5 per primer. For the $30 for the 6 primers, the cost per test is $0.06, and it gets much cheaper with bigger orders!
The last thing we need is BST polymerase, which can be purchased online. The reaction needs about 8 'units' of material, and 8000 units cost $283.00. That's about $0.28 per test and is the most expensive component for the test. Of course, you can make your own too, but that's a bit beyond the scope of this project. Next time!
Protocol
To perform a COVID-19 test, you just need to mix the materials described above as describe in one of the protocols available online, into a PCR tube.
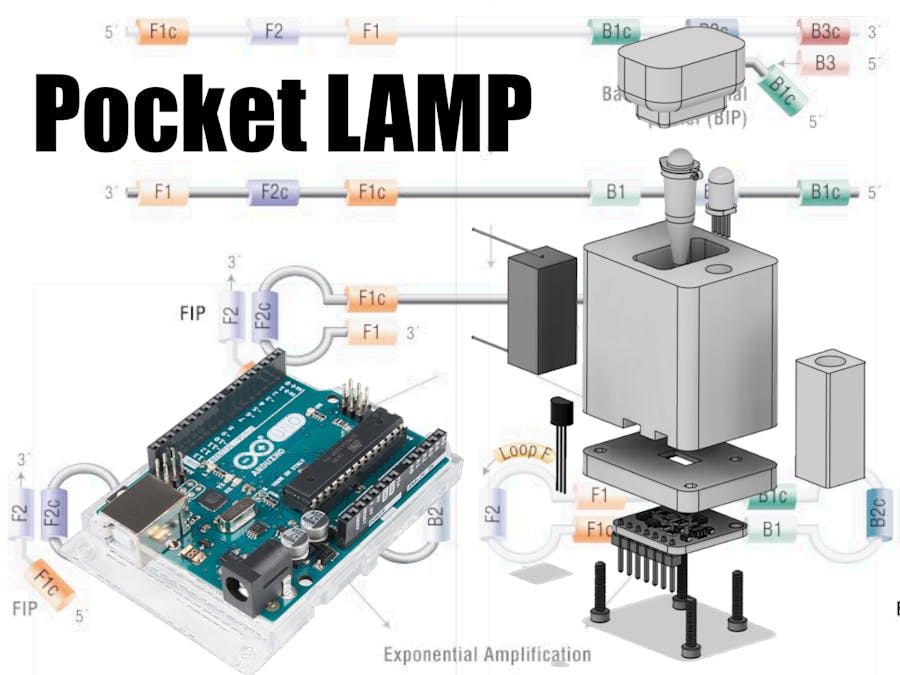
ElectronicsTo build our device, we are going to need various circuits to control, measure, and analyze the reaction. The goal here is a cheap and rapid prototype, so we will use off-the-shelf parts you can find in most maker-spaces.
But what's more important than cheap and rapid here is safe!
So from the beginning, we will be focusing on how we can detect possible sources of error that would give an inaccurate result.
The control circuitry will use an Arduino, as they are easy to code and obtain, and we will use it for user and temperature control, and interfacing our optical sensors.
Optical Detection
What we basically need is a spectrofluorophotometer to detect the color of the reaction mixture. A super cheap and simple solution here is the TCS34725 breakout board, with a simple modification. We can use each of the red, green, and blue channels to determine the state of the device, and be sure everything is operating correctly!
In the most basic analysis, each color channel can measure a high or low level of light, lets quickly go through the combinations:
- Red: High, Blue: either, Green: either = Lid is open!
- Red: low, Blue: low, Green: either = Lid Closed, UV LED is not on.
- Red: low, Blue: high, Green: low = Lid Closed, UV LED on, Nothing detected yet
- Red: low, Blue: high, Green: high = Lid Closed, UV LED on, LAMP has detected something!
So, we can use the extra color channels to detect user error (they forgot to close the lid, so the sample didn't heat correctly), or device failure (UV LED is broken), both of which could lead to false negatives! A false negative is much worse than a false positive, as someone is inconvenienced by a follow-up test is less dangerous than a sick person missing out on treatment and spreading the disease.
The fluorescence spectrum of Calcein looks like this:
The bar at the top is the approximate color our eyes see, but a lot is in the ultraviolet or infrared. The dotted show the range and effect of incoming light, with 'excitation' in blue, and 'emission' in green. On the excitation spectra, the higher the curve the better the calcein is at absorbing light. On the emission spectra, the higher the curve, the more likely a photon 'emitting' from the calcein will have this color. It very quantum, and something for another project to describe (I'm hoping to build a Raman spectroscope one day).
We will use the VLMU3100 UV LED to excite the calcein. Although it's not at the very optimal range to excite the dye, it has less overlap with the green sensor in the TCS34725 color sensor than a blue LED would produce. That way we can better separate the LED signal from the LAMP signal,
For our initial tests, we can use a sample of the compound Fluorescein (similar to Calcein optically, but not sensitive to Magnesium or Manganese). We just need to show that the wavelength and intensity of this LED are sufficient for a 20 mMol sample, which it is! The sample in the top picture is pure water, and the UV light from the LED has no visible effect. In the bottom picture, we have a tiny sample of fluorescein in water, at a concentration on 20 micromoles. Under UV light, the pale yellow solution glows bright green!
We want to replace the white LED on the TCS34725 with the UV VLMU3100, so we will also have to replace the current-limiting resistor. As the forward drop on the UV LED is 3.2 V, and we want about 15 mA through the diode, we will replace the 33 Ohm SMD resistor with a 6.8 Ohm one (which, against all odds, I happen to have in my SMD resistor tray!).
Carefully replacing both parts, we can test the UV LED. In the picture below it glows blue, but as cameras can't record far blue light, it's nowhere near as bright as seen live!
Connecting everything to the Arduino Uno, we can now control the UV LED programmatically, and record Red-Green-Blue_white light levels.
Temperature Control
The LAMP reaction requires a very steady temperature to be maintained throughout the reaction. The reactions will be done in tiny containers called PCR tubes. They hold 50 microliters of fluid, about the volume of a drop of water. The walls of the container are very thin, and allow the fluid within to be heated quickly.
For heating the liquid to the correct temperature, the tube will be inserted into a hole in a block of aluminum. The block will be heated by a wire wound resistor and monitored by a MAX31820 1-wire temperature sensor. The heating assembly will be controlled by the Arduino using a PID loop.
The aluminum heat block is simply a piece of square rod cut and drilled out by hand. The material is cheap and easy to find.
The assembled heating unit looks like this, before going into the housing. Its held together by a few small blogs of epoxy heat paste.
Designing the Case
It is important that the Pocket Lamp can be built where it is needed, so I have designed a housing that can be printed using less than 20 grams of PLA filament.
Each component has one flat surface, so build adhesion is great! I have included Fusion 360 and STL files in the repo.
Assembling
Once the parts are printed, the electronic components fit together with the aluminum heat block and are held together with 12 mm M2 bolts.
The tri-color LED has the correct resistors soldered on and slides into place inside the housing (220, 100, 100 Ohms for R, G, and B respectively), The glued-together heating system also slides in easily, and the TCS34725 and base plate holds everything together.
FreeStyle Soldering
A bent pin-header strip is used to join the remaining electronic components, and connect to the Arduino Uno. The next iteration with of course use a custom PCB, but I really like the look of freestyle electronics :)
All that's left is to plug the device into your Arduino, and you have an amazing, virus-detecting Shield!
As most of the amazing stuff is going on in the PCR tube, our job is simply to monitor what's going on and output the results. This is usually done in a tremendously expensive device ($$$ 5-7 figures), but fundamentally all we need to do is the following:
1) Calibrate the Fluorescence Spectroscope
2) Warm the sample holder to the correct temperature for the LAMP reaction (59 Celcius, using a PID loop)
3) Alert the user to insert the sample, and start the timer. The sample temperature has to be kept very steady!
4) Stream the data (intensity/wavelength, temperature, time packets) over serial for logging. One data point per second is enough.
5) Report the results for the COVID-19 test (Error, Negative, Positive)
A professional machine can run hundreds or thousands of samples simultaneously and has incredibly accurate temperature control. But we can do all the fundamental operations on an Arduino. And luckily, there are Arduino libraries for all the components and PID-loop required! We can just glue these together and have our device ready quickly!
The last thing we have to consider is our user interface. We have one RGB LED for notifications. Let's define our User Interface and stages:
Warming up: Slow Blue color cycling (1 Hz). The lid should be on, and we can calibrate the UV LED brightness now.
Ready to Starts, Please insert sample: Solid Green. We can use the changes in brightness to see when the user has opened and closed the lid, no switch necessary :)
Running the Sample: Blinking Green LED
Error Detected: Blinking Red LED
Results:
Cyan: Nothing Detected.
Purple: Positive LAMP result, time for follow-up tests!
After the test procedure is complete, the user just resets the device to performs a new test!
If the Pocket Lamp operator has a PC (it's only necessary to use one to program the Arduino, from then on it's stand-alone), they can capture the streamed data on the serial port for further processing.
The code is available on GitHub, along with the 3D files, documentation on the components, and circuit schematics.
ConclusionThe COVID-19 crisis needs new models for getting testing and medicine to the entire world. One such model is a distributed system, where cheap devices and technology gets spread to where it's needed.
It won't ever be as cost-effective as a centralized approach, with industrial-scale testing machines running 24/7. But, in countries where there aren't large amounts of capital to invest in expensive equipment and personal, or the country's transport infrastructure just can't get samples to a centralized point quickly or efficiently enough, a distributed system might be the only way.
The laboratory technology for COVID-19 testing has developed amazingly rapidly in the past few months. That technology needs to be packed into systems that can make it out of the lab and into people's hands around the globe. I hope this project has demonstrated that even the most cutting-edge technology can sometimes be put in the hands of Makers!




_ztBMuBhMHo.jpg?auto=compress%2Cformat&w=48&h=48&fit=fill&bg=ffffff)



Comments